Histone Modifications Marks
H3K4me3 and/or H3K27ac
Ultra-Low Sample Input
Requires 1-2 million cells and offers
2x sensitivity
Nano ChIP-Seq and RNA-Seq
Signomax™ is a comprehensive sample-to-report solution that integrates RNA Sequencing (RNA-Seq) and/or Nano-ChIP Sequencing (Nano-ChIP-Seq) with proprietary, customizable bioinformatics analysis. Designed to empower researchers, Signomax™ delivers publication-ready data and unique epigenomic insights to support retrospective studies, cohort analyses, and biomarker discovery.
We offer RNA-Seq services for Formalin-Fixed Paraffin-Embedded (FFPE) samples. Our library
preparation service accommodates low sample input and places no limits on sample quality (DV200).

Table 1: Comparison of sample requirements between Auristone and other vendors
Compared to Vendor A’s RNA-Seq library preparation on FFPE samples, Auristone delivers a
consistent yield exceeding tenfold, while maintaining a higher pass rate*.
*Pass rate refers to libraries that pass Quality Check (QC) metrics and requirements for sequencing
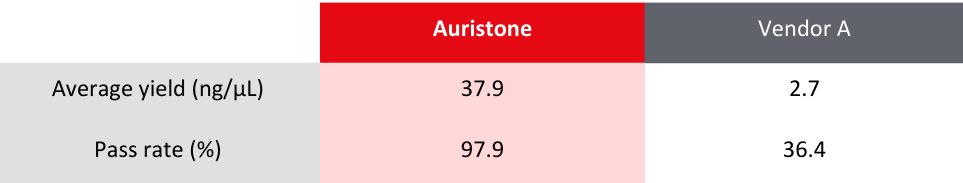
Table 2: Comparison of average yield and pass rate for library preparation between Auristone and Vendor A
We provide RNA-Seq data processing and analysis, including differential gene expression, gene set enrichment analysis and specific signature visualizations, among other analyses.
We are dedicated to supporting your research endeavours and accelerating your scientific journey. Our aim is to help you achieve your goals efficiently. With a focus on excellence, we offer fast turnaround times, ensuring you receive publication-ready data promptly. Should you have any additional requirements or questions, please feel free to contact us at enquiry@auristone.com.
Auristone employs a proprietary Nano-scaled Chromatin Immunoprecipitation Sequencing (Nano ChIP-Seq) workflow to explore post-translational histone modifications in samples. ChIP-Seq is a powerful technique that integrates ChIP assays with Next-Generation Sequencing (NGS) to examine interactions between DNA and proteins. The nano-scaled nature of our ChIP-Seq protocol stems from the requirement for ultra-low sample input. This ultra-sensitive technology enables a wider-range of applications for discovery, validation, and screening of genome-wide histone modifications in a reliable and efficient manner. With these capabilities, Signomax™ effectively addresses unmet needs in epigenomic biomarker discovery.
H3K4me3 and/or H3K27ac
Requires 1-2 million cells and offers
2x sensitivity
Accepts cell lines, fresh frozen and
Formalin-Fixed Paraffin-Embedded
(FFPE) slides
Developed a streamlined and highly
reproducible workflow from sample
preparation to data analysis
Specific antibody is used to precipitate DNA-bound Protein of Interest (POI)
DNA bound to the POI is isolated
Isolated DNA fragments are sequenced to determine DNA binding sites for POI
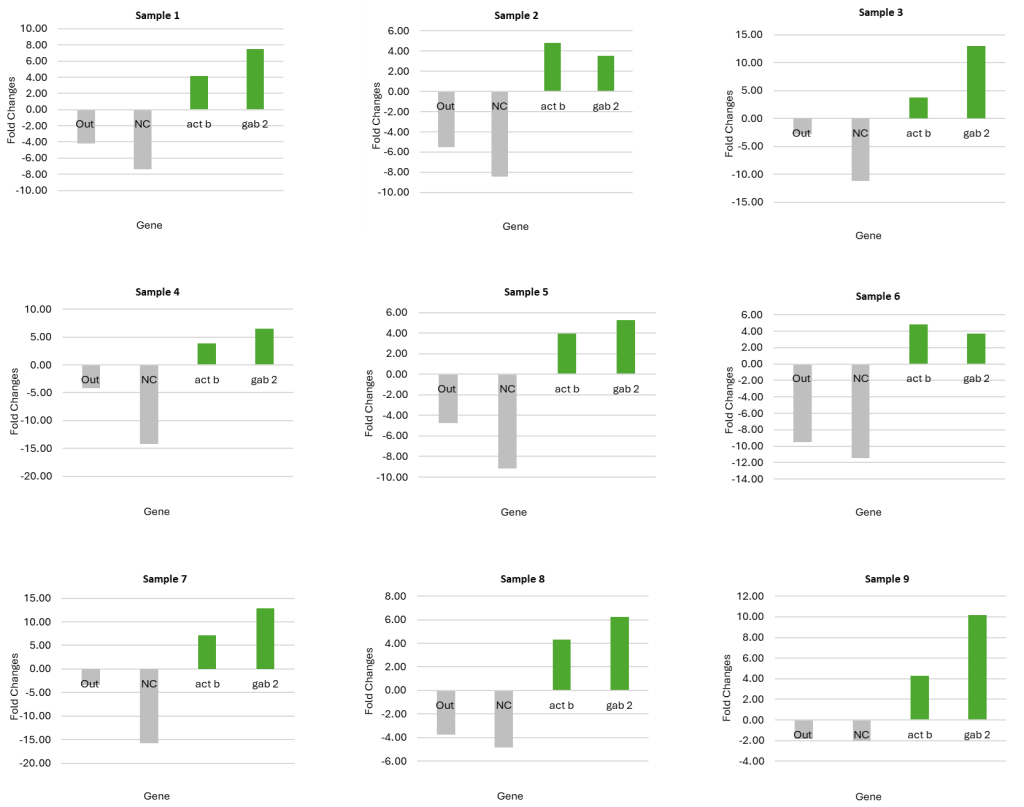
Fig 1. Graphical representation of ChIP-Seq qPCR quality control data, illustrating fold change between ChIP-enriched DNA and input controls. This metric validates successful target enrichment before proceeding to next generation sequencing.
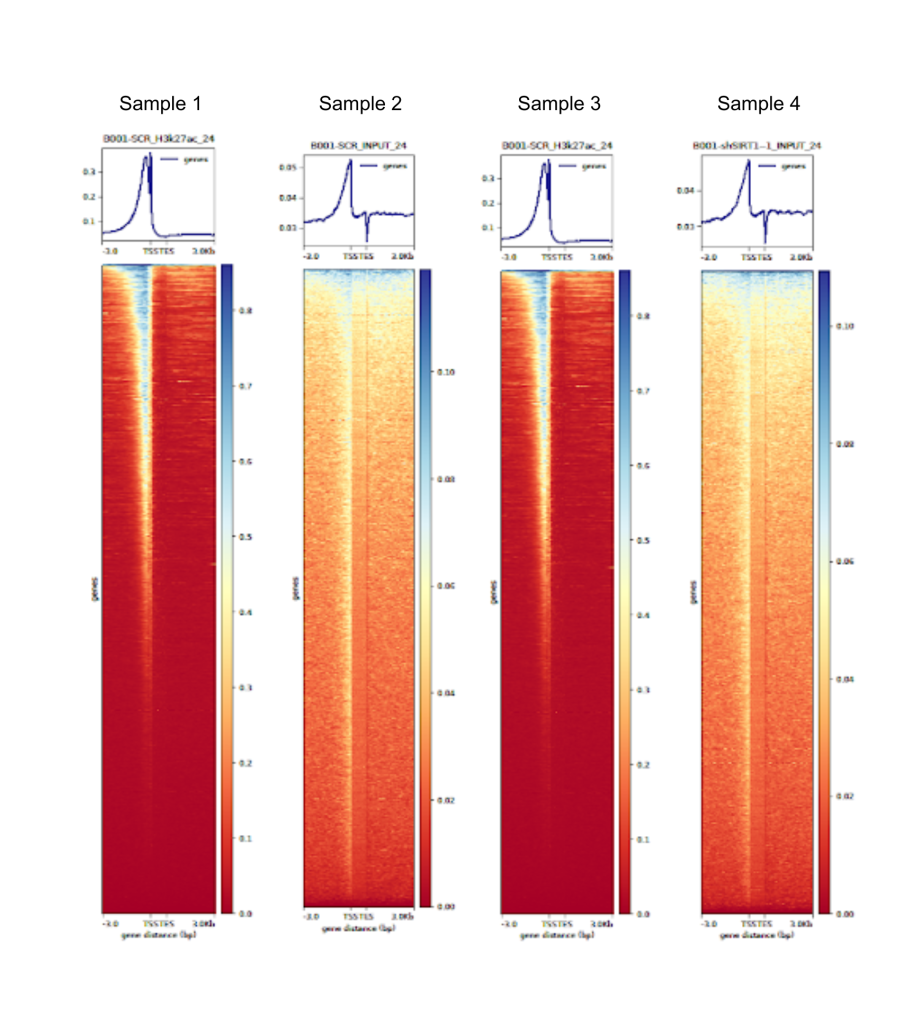
Fig 2. Generated through the Signomax™ pipeline, this heatmap compares four distinct ChIP-Seq samples, highlighting differences and similarities in their genomic enrichment patterns.
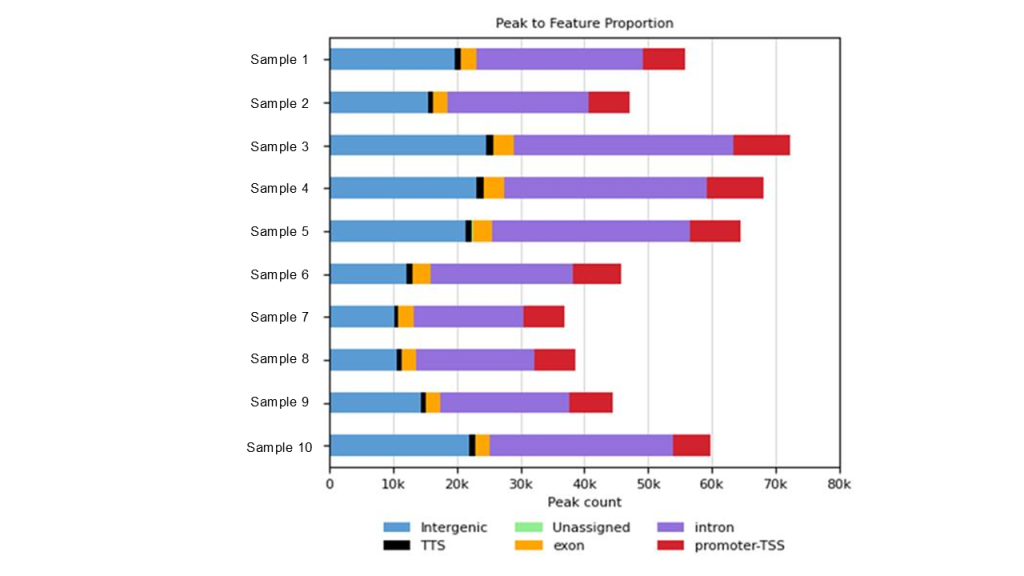
Fig 3. Visualization of ChIP-Seq peak distribution across key genomic features in 10 samples, processed through the Signomax™ pipeline. This breakdown highlights promoter and enhancer enrichments critical for functional genomics.
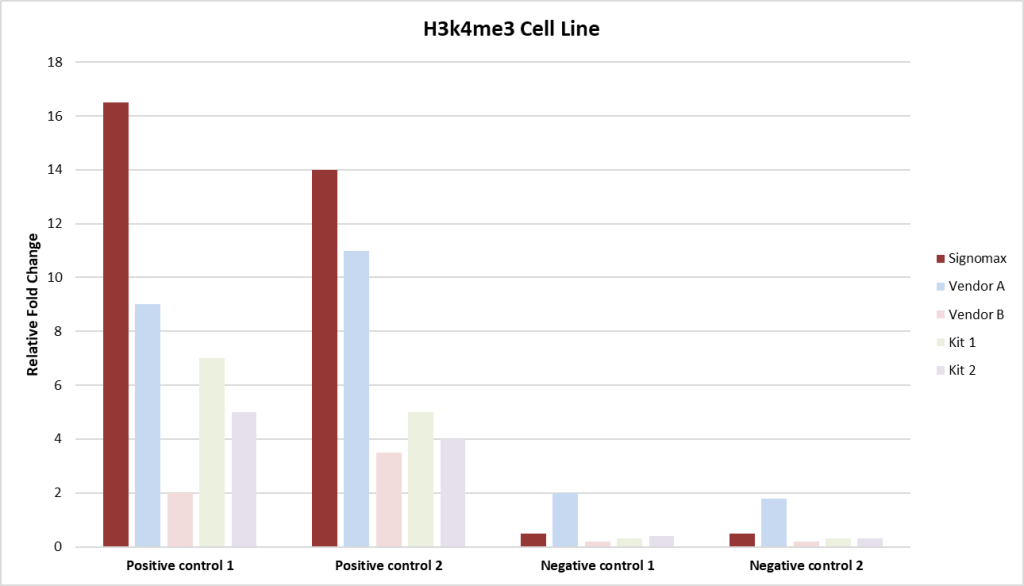
Fig 4. This chart displays qPCR fold change comparisons between Signomax™ and other commercial ChIP-Seq kits and service providers. Signomax™ shows consistently higher fold change, indicating enhanced target enrichment and assay sensitivity.
For more information on Signomax™, please contact enquiry@auristone.com
Interested in working with us? Drop us a note below and we’ll get back to you as soon as we can.
enquiry@auristone.com
LaunchPad @ one-north
77 Ayer Rajah Crescent #01-27 Singapore 139954